
The applied phylogenetics lab at usc
Our team uses phylogenetic trees, graphical depictions of historical relationships, to study evolution at multiple scales—from the grandest (e.g., the origin of major groups of organisms) to the smallest (e.g., the development of the adaptive immune system within an individual) of scales. In our research, we develop computational and statistical tools, analyze mathematical models, and conduct large-scale empirical analyses to answer evolutionary questions.
Macroevolution and the genotype-phenotype map
We work to understand the origins of phenotypic diversity and the evolution of the genetic architecture that underlies it.

Evolutionary immunology
We use phylogenetic approaches to understand the evolutionary dynamics of antibodies and the genes that encode them.
Lineage diversification
We seek general explanations for why some lineages are so much more diverse than others and to characterize how much we can learn from phylogenetic data about this phenomenon.

Our Team
We’re a diverse group of people, with a diverse set of backgrounds, interests, and expertise. We are united by a shared passion for applying rigorous quantitative reasoning and evolutionary principles to understand biological phenomena.
We are located in the University of Southern California’s Department of Quantitative and Computational Biology, the oldest, and one of the most recognized, computational biology groups in the US. And we live, work, and play in Los Angeles, the most interesting city in the world
Interested in joining the lab?
We are always on the lookout for smart, ambitious people to join our team. If you’re interested in working alongside us on cool problems, please get in touch.
Latest Publications

The evolutionary and functional significance of germline immunoglobulin gene variation
Recent research has revealed a great deal of previously undocumented variation among individuals in their immunoglobulin genes — the part of the genome that codes for the antigen-recognition sequence of antibodies. In a new paper, we review findings from multiple lines of inquiry that may reveal the evolutionary processes that maintain this variation.
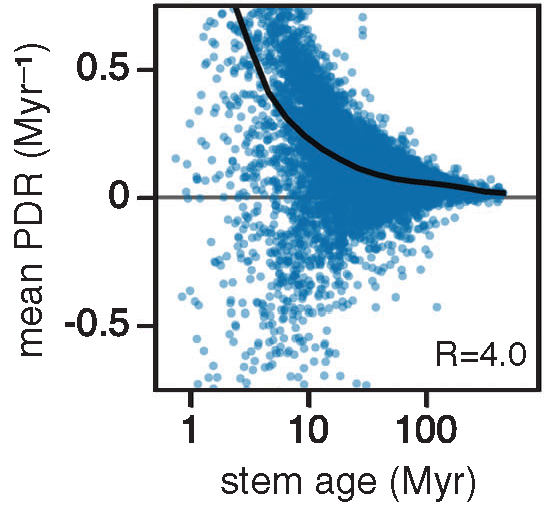
The scaling of diversification rates with age is likely explained by sampling bias
Many empirical analyses of diversification rates (including a previous study from our lab, led by Francisco Henao Diaz) have found that they scale with age, with younger taxa seemingly diversifying faster. In a new paper, we demonstrate that this phenomenon is likely explained by a sampling bias.
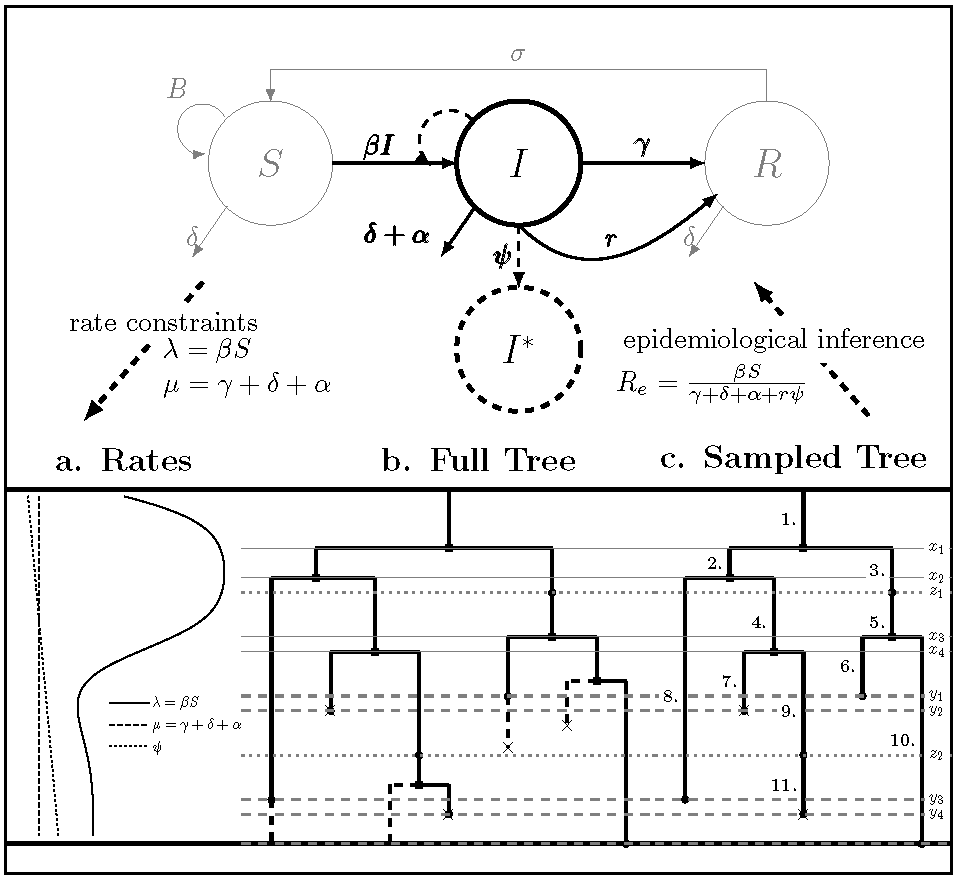
A General Birth-Death-Sampling Model for Epidemiology and Macroevolution
There are a multitude of birth-death phylogenetic models used in macroevolution and epidemiology. In a paper led by Ailene MacPherson, we proved that all of these are actually variants of a more general model and provided a new technique for deriving likelihoods for any submodel.



The evolutionary and functional significance of germline immunoglobulin gene variation
The scaling of diversification rates with age is likely explained by sampling bias
A General Birth-Death-Sampling Model for Epidemiology and Macroevolution
Recent research has revealed a great deal of previously undocumented variation among individuals in their immunoglobulin genes — the part of the genome that codes for the antigen-recognition sequence of antibodies. In a new paper, we review findings from multiple lines of inquiry that may reveal the evolutionary processes that maintain this variation.
Many empirical analyses of diversification rates (including a previous study from our lab, led by Francisco Henao Diaz) have found that they scale with age, with younger taxa seemingly diversifying faster. In a new paper, we demonstrate that this phenomenon is likely explained by a sampling bias.
There are a multitude of birth-death phylogenetic models used in macroevolution and epidemiology. In a paper led by Ailene MacPherson, we proved that all of these are actually variants of a more general model and provided a new technique for deriving likelihoods for any submodel.

